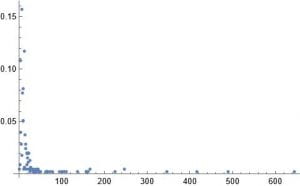
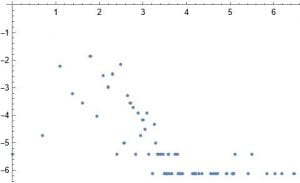
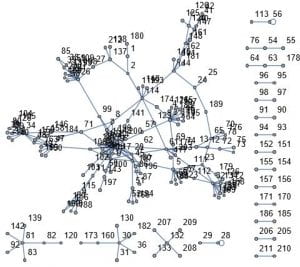
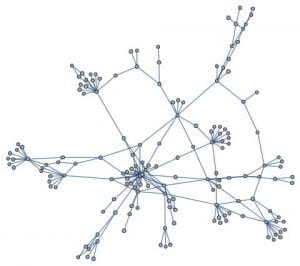
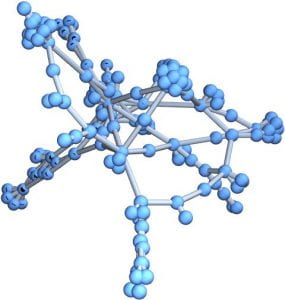
This week I decided to work on a random duplication network. I start with a graph of a ppi network 1 – 20 vertices. I then pick at random a vertex to duplicate. In my case the random choice was 12. I then join this vertex to 11 and 13. I add these new edges to the original network. I do this many times. After doing this 160 times this is the result.